Fold change (\(\Delta \Delta C_T\) method) analysis of repeated measure qPCR data
Source:R/qpcrREPEATED.r
qpcrREPEATED.RdqpcrREPEATED function performs fold change (\(\Delta \Delta C_T\) method)
analysis of observations repeatedly taken over different time courses.
Data may be obtained over time from a uni- or multi-factorial experiment. The bar plot of the fold changes (FC)
values along with the standard error (se) or confidence interval (ci) is also returned by the qpcrREPEATED function.
qpcrREPEATED(
x,
numberOfrefGenes,
factor,
block,
width = 0.5,
fill = "#BFEFFF",
y.axis.adjust = 2,
y.axis.by = 1,
ylab = "Fold Change",
xlab = "none",
fontsize = 12,
fontsizePvalue = 7,
axis.text.x.angle = 0,
axis.text.x.hjust = 0.5,
x.axis.labels.rename = "none",
letter.position.adjust = 0,
p.adj = "none",
errorbar = "se",
plot = TRUE
)Arguments
- x
input data frame in which the first column is id, followed by the factor column(s) which include at least time. The first level of time in data frame is used as calibrator or reference level. Additional factor(s) may also be present. Other columns are efficiency and Ct values of target and reference genes. NOTE: In the "id" column, a unique number is assigned to each individual from which samples have been taken over time, for example see
data_repeated_measure_1, all the three number 1 indicate one individual which has been sampled over three different time courses. Seevignette, section "data structure and column arrangement" for details.- numberOfrefGenes
number of reference genes which is 1 or 2 (Up to two reference genes can be handled). as reference or calibrator which is the reference level or sample that all others are compared to. Examples are untreated of time 0. The FC value of the reference or calibrator level is 1 because it is not changed compared to itself. If NULL, the first level of the main factor column is used as calibrator.
- factor
the factor for which the FC values is analysed. The first level of the specified factor in the input data frame is used as calibrator.
- block
column name of the block if there is a blocking factor (for correct column arrangement see example data.). Block effect is usually considered as random and its interaction with any main effect is not considered.
- width
a positive number determining bar width in the output bar plot.
- fill
specify the fill color for the columns in the bar plot. If a vector of two colors is specified, the reference level is differentialy colored.
- y.axis.adjust
a negative or positive value for reducing or increasing the length of the y axis.
- y.axis.by
determines y axis step length
- ylab
the title of the y axis
- xlab
the title of the x axis
- fontsize
font size of the plot
- fontsizePvalue
font size of the pvalue labels
- axis.text.x.angle
angle of x axis text
- axis.text.x.hjust
horizontal justification of x axis text
- x.axis.labels.rename
a vector replacing the x axis labels in the bar plot
- letter.position.adjust
adjust the distance between the signs and the error bars.
- p.adj
Method for adjusting p values
- errorbar
Type of error bar, can be
seorci.- plot
if
FALSE, prevents the plot.
Value
A list with 5 elements:
- Final_data
Input data frame plus the weighted Delat Ct values (wDCt)
- lm
lm of factorial analysis-tyle
- ANOVA_table
ANOVA table
- FC Table
Table of FC values, significance, confidence interval and standard error with the lower and upper limits for the selected factor levels.
- Bar plot of FC values
Bar plot of the fold change values for the main factor levels.
Details
The qpcrREPEATED function performs fold change (FC) analysis of observations repeatedly taken over time.
The intended factor (could be time or any other factor) is defined for the analysis by the factor argument,
then the function performs FC analysis on its levels
so that the first levels (as appears in the input data frame) is used as reference or calibrator.
The function returns FC values along with confidence interval and standard error for the FC values.
Examples
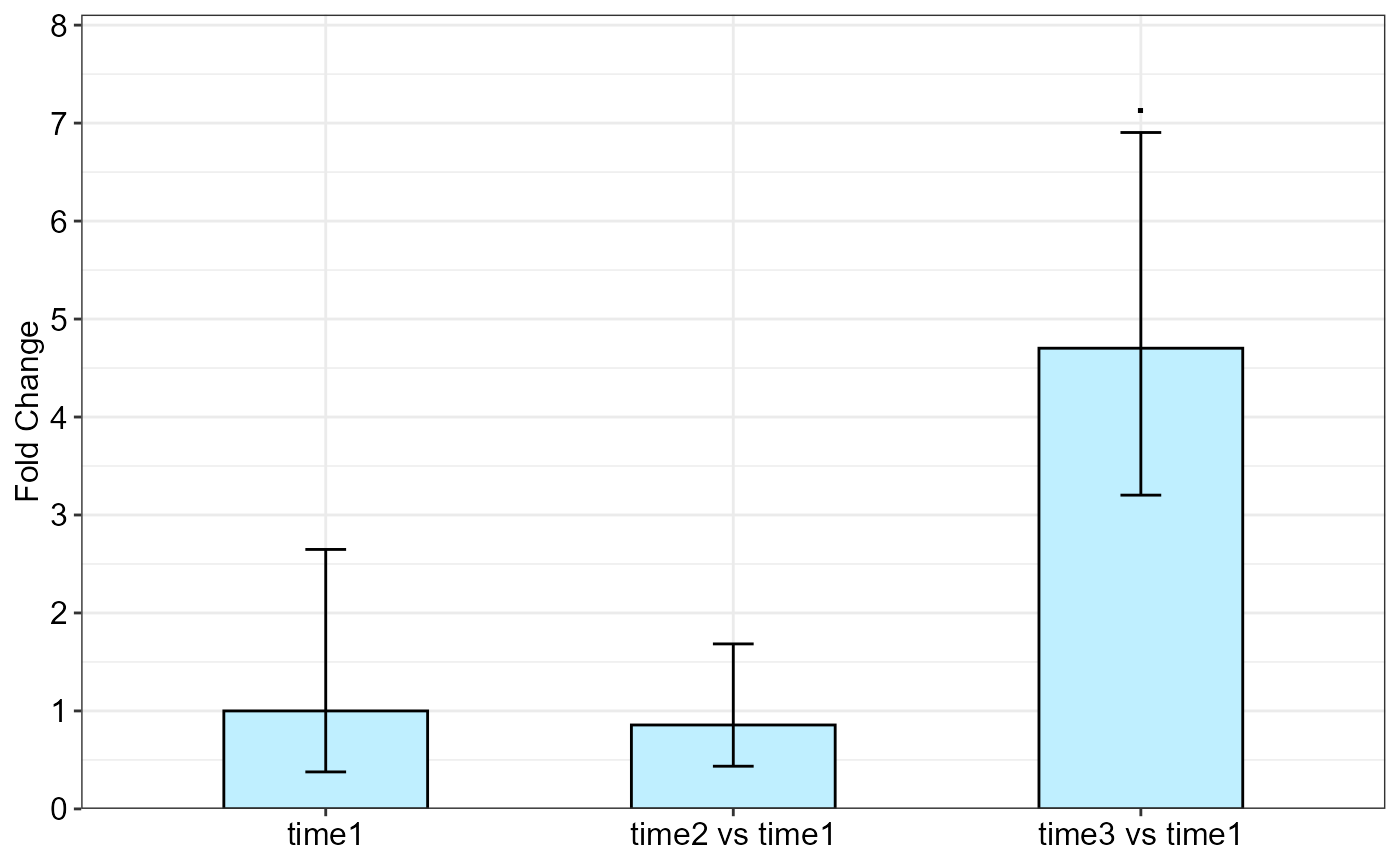
qpcrREPEATED(data_repeated_measure_1,
numberOfrefGenes = 1,
factor = "time", block = NULL)
#> Warning: The level 1 of the selected factor was used as calibrator.
#> Type III Analysis of Variance Table with Satterthwaite's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> time 11.073 5.5364 2 4 4.5382 0.09357 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Fold Change table
#> contrast FC pvalue sig LCL UCL se Lower.se Upper.se
#> 1 time1 1.0000 1.0000 0.0000 0.0000 1.4051 0.3776 2.6484
#> 2 time2 vs time1 0.8566 0.8166 0.0923 7.9492 0.9753 0.4357 1.6841
#> 3 time3 vs time1 4.7022 0.0685 . 0.5067 43.6368 0.5541 3.2026 6.9040
#>
#> Fold Change plot of the main factor levels
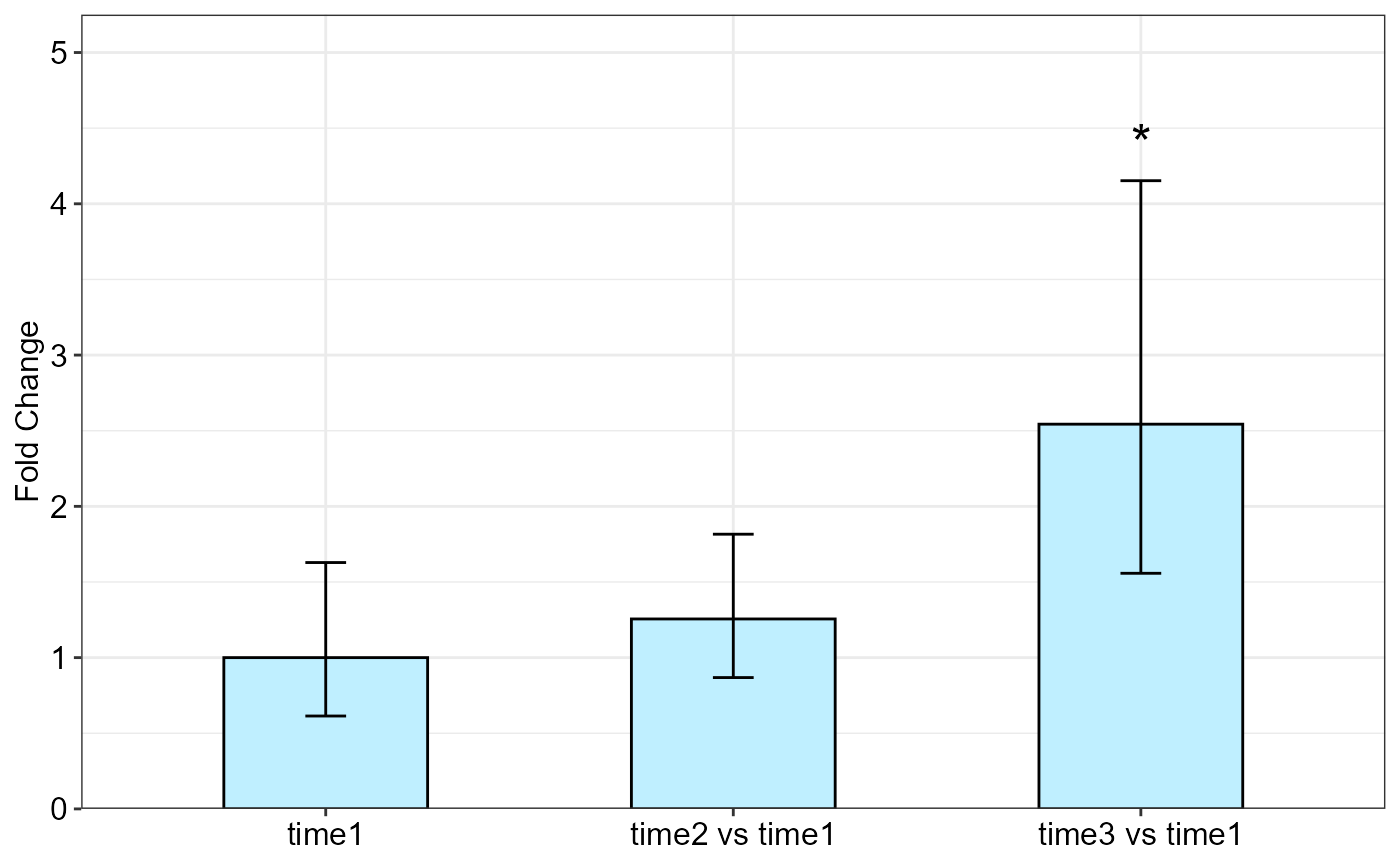
 qpcrREPEATED(data_repeated_measure_2,
numberOfrefGenes = 1,
factor = "time", block = NULL)
#> Warning: The level 1 of the selected factor was used as calibrator.
#> NOTE: Results may be misleading due to involvement in interactions
#> Type III Analysis of Variance Table with Satterthwaite's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> time 5.9166 2.95832 2 8 3.4888 0.08139 .
#> treatment 0.6773 0.67728 1 4 0.7987 0.42199
#> time:treatment 6.3186 3.15932 2 8 3.7258 0.07186 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Fold Change table
#> contrast FC pvalue sig LCL UCL se Lower.se Upper.se
#> 1 time1 1.0000 1.0000 0.0000 0.0000 0.7036 0.6140 1.6286
#> 2 time2 vs time1 1.2556 0.5540 0.4381 3.5987 0.5323 0.8682 1.8159
#> 3 time3 vs time1 2.5432 0.0351 * 0.8873 7.2895 0.7074 1.5575 4.1527
#>
#> Fold Change plot of the main factor levels
qpcrREPEATED(data_repeated_measure_2,
numberOfrefGenes = 1,
factor = "time", block = NULL)
#> Warning: The level 1 of the selected factor was used as calibrator.
#> NOTE: Results may be misleading due to involvement in interactions
#> Type III Analysis of Variance Table with Satterthwaite's method
#> Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
#> time 5.9166 2.95832 2 8 3.4888 0.08139 .
#> treatment 0.6773 0.67728 1 4 0.7987 0.42199
#> time:treatment 6.3186 3.15932 2 8 3.7258 0.07186 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Fold Change table
#> contrast FC pvalue sig LCL UCL se Lower.se Upper.se
#> 1 time1 1.0000 1.0000 0.0000 0.0000 0.7036 0.6140 1.6286
#> 2 time2 vs time1 1.2556 0.5540 0.4381 3.5987 0.5323 0.8682 1.8159
#> 3 time3 vs time1 2.5432 0.0351 * 0.8873 7.2895 0.7074 1.5575 4.1527
#>
#> Fold Change plot of the main factor levels