The efficiency function calculates amplification efficiency and returns related statistics and standard curves.
efficiency(df)Arguments
- df
a data frame of dilutions and Ct of genes. First column is dilutions and other columns are Ct values for different genes.
Value
A list 3 elements.
- efficiency
Slope, R2 and Efficiency (E) statistics
- Slope_compare
slope comparison table
- plot
standard curves
Details
The efficiency function calculates amplification efficiency of genes, and present the Slope, Efficiency, and R2 statistics.
Examples
# locate and read the sample data
data_efficiency
#> dilutions C2H2.26 C2H2.01 GAPDH
#> 1 1.00 25.57823 24.25 22.60794
#> 2 1.00 25.53636 24.13 22.68348
#> 3 1.00 25.50280 24.04 22.62602
#> 4 0.50 26.70615 25.56 23.67162
#> 5 0.50 26.72720 25.43 23.64855
#> 6 0.50 26.86921 26.01 23.70494
#> 7 0.20 28.16874 27.37 25.11064
#> 8 0.20 28.06759 26.94 25.11985
#> 9 0.20 28.10531 27.14 25.10976
#> 10 0.10 29.19743 28.05 26.16919
#> 11 0.10 29.49406 28.89 26.15119
#> 12 0.10 29.07117 28.32 26.15019
#> 13 0.05 30.16878 29.50 27.11533
#> 14 0.05 30.14193 29.93 27.13934
#> 15 0.05 30.11671 29.71 27.16338
#> 16 0.02 31.34969 30.69 28.52016
#> 17 0.02 31.35254 30.54 28.57228
#> 18 0.02 31.34804 30.04 28.53100
#> 19 0.01 32.55013 31.12 29.49048
#> 20 0.01 32.45329 31.29 29.48433
#> 21 0.01 32.27515 31.15 29.26234
# Applying the efficiency function
efficiency(data_efficiency)
#> $Efficiency
#> Gene Slope R2 E
#> 1 C2H2.26 -3.388094 0.9965504 1.973110
#> 2 C2H2.01 -3.528125 0.9713914 1.920599
#> 3 GAPDH -3.414551 0.9990278 1.962747
#>
#> $Slope_compare
#> $emtrends
#> variable log10(dilutions).trend SE df lower.CL upper.CL
#> C2H2.26 -3.39 0.0856 57 -3.56 -3.22
#> C2H2.01 -3.53 0.0856 57 -3.70 -3.36
#> GAPDH -3.41 0.0856 57 -3.59 -3.24
#>
#> Confidence level used: 0.95
#>
#> $contrasts
#> contrast estimate SE df t.ratio p.value
#> C2H2.26 - C2H2.01 0.1400 0.121 57 1.157 0.4837
#> C2H2.26 - GAPDH 0.0265 0.121 57 0.219 0.9740
#> C2H2.01 - GAPDH -0.1136 0.121 57 -0.938 0.6186
#>
#> P value adjustment: tukey method for comparing a family of 3 estimates
#>
#>
#> $plot
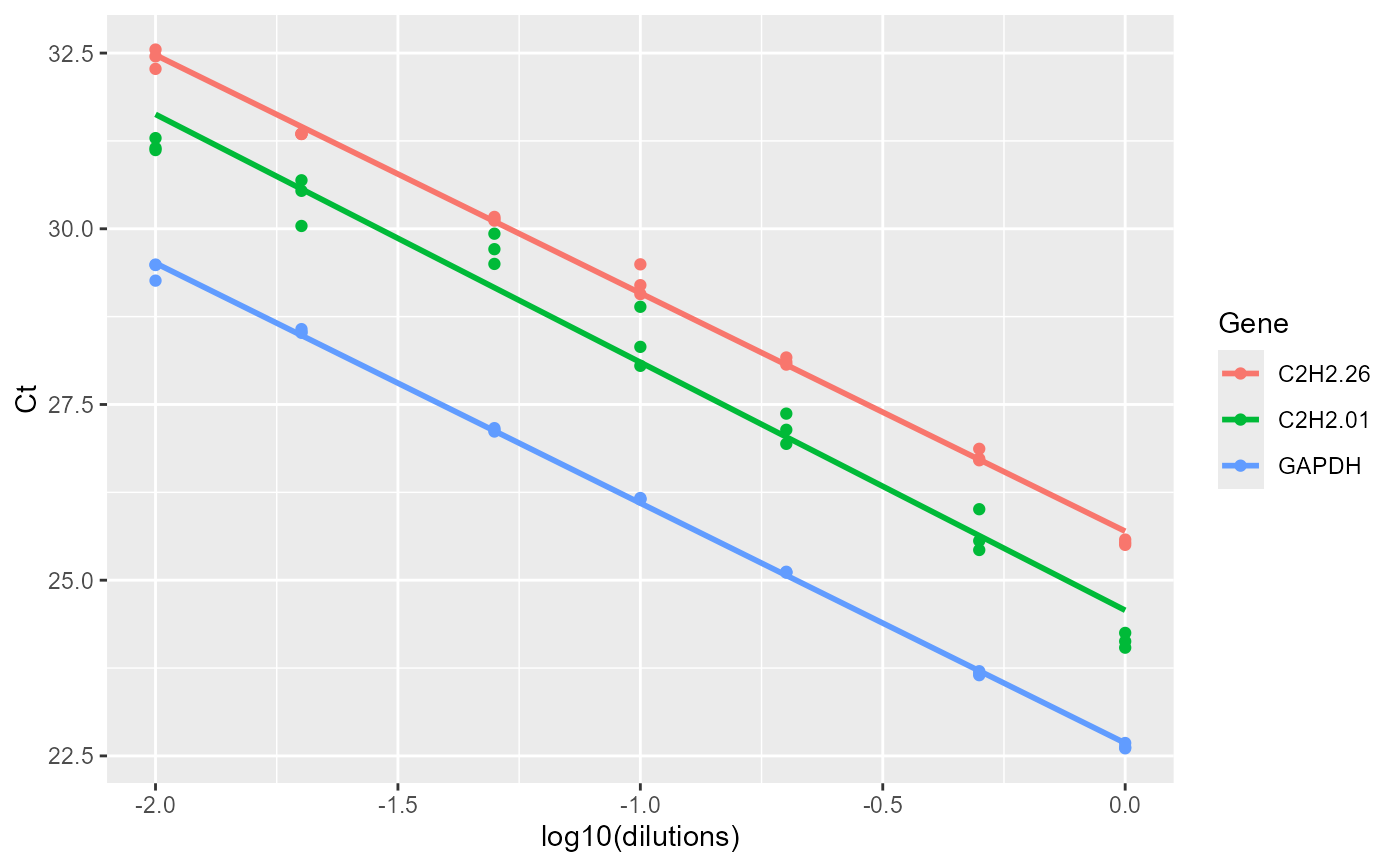 #>
#>